It’s not ALL in the genes—the role of epigenetics
Expert reviewers
Essentials
- Epigenetics is an additional layer of instructions that controls how our DNA is interpreted—how our genes are controlled and expressed
- Epigenetic mechanisms change the way genes are packaged in the cell nucleus, and involve changes in chemical groups that can attach to DNA, or changes in the way RNA molecules interact with our DNA
- Some epigenetic mechanisms can go wrong during development or cell division, resulting in disease
- Diet and other external environmental influences can potentially play a role in controlling epigenetic processes
- Some epigenetic marks can potentially be passed down through the generations
We’re pretty used to thinking ‘it’s all in the genes’… that we are who we are because of the genes we received from our parents. But what if we are who we are not only because of the genes our parents gave us, but also because of the environment in which our grandparents lived? Turns out, it’s not ALL in the genes—external and environmental factors can influence the way an organism’s genes are regulated. This is just part of a field of genetics known as epigenetics—‘epi’ coming from the Greek for ‘over, on top of’. Epigenetics is an additional layer of instructions that lies ‘on top of’ DNA, controlling how the genes are read and expressed.
Pretty much every cell of an organism, be it a parrot, a panther, a plum tree or a person, contains the complete set of instructions to build that organism—its DNA. DNA is a very long molecule, made up of smaller pieces known as nucleotide bases. Humans have 3 billion nucleotide bases in their DNA. The sequence of these bases is ever so slightly different between individuals, which is the key to our being unique.
Put certain combinations of these bases together, and you get genes, which provide the information that determines particular characteristics. So, the DNA that is contained in the nucleus of nearly every cell of our body contains all of our genes, plus much more extra DNA that is required to ensure the genes are properly controlled. Collectively, this is known as our genome.
But hang on, if every cell contains ALL the DNA and the DNA contains ALL the genes and ALL the instructions, how do we end up with all the different cells that make up all the different body parts? We obviously are not made purely of brain cells—we also need heart cells, skin cells, liver cells and lots more.
So there’s got to be some sort of control mechanism that ‘regulates’ all these genes in the DNA in our various cells—a mechanism that can turn some genes on, but keep others quiet in order to build functional cells, that can go on to build organs and tissues. This process is called ‘cell differentiation’ and is just one example, indeed, a classic example, of epigenetics—there’s an extra layer of information lying above the actual genes and surrounding DNA, telling them which ones to switch on or off to build each of the different functional cells our bodies need.
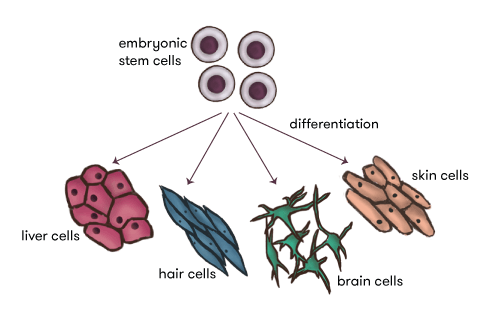
Epigenetics can be thought of as the interpretation of the genetic code. Just as the same piece of music will change slightly when interpreted by different orchestras, so does our genetic ‘score’ when interpreted by the epigenetic orchestra.
Epigenetic mechanisms
A few basic definitions
Before we get into the guts of epigenetics, it’s useful to go through a few quick definitions of terms that are going to keep cropping up.
- DNA
- A long molecule that is made up of nucleotide bases, carbon sugar molecules and phosphate molecules. To make the twisted ladder shape of the DNA double helix, the nucleotide bases pair up to make the rungs, with the sugar and phosphate molecules along the sides. There are four nucleotide bases: adenine, thymine, guanine and cytosine.
- RNA
- A single stranded molecule that is copied from DNA and also made of nucleotide bases, sugar molecules and phosphate groups. There are several different types of RNA that each have a different function.
- messenger RNA
- A specific class of RNA strand that is used as a carrier of the information required to build proteins from DNA.
- gene
- A distinct section of DNA that provides the information that is copied into messenger RNA to build particular proteins, that then go on to build specific cell types.
- genome
- The entirety of an organism’s DNA—the DNA that codes for genes, and the extra (non-coding) DNA that is used to provide landmarks which regulate which genes are switched on or off.
- transcription
- The process through which the DNA sequence of genes is read to produce messenger RNA, which then uses it as a template to build proteins.
Back to the actual epigenetics. There are a few different ways epigenetic instructions can be ‘recorded’ in the genome: histone modification, DNA methylation and the actions of different types of RNA.
Histone modification
The DNA within the cell nuclei is not just one super-long molecule all tangled up like a ball of string. It’s very neatly organised; wrapped around molecules called histones to make structures called nucleosomes. Nucleosomes are kind of like beads on a necklace that can be either strung very tightly together, or spaced far apart. This spacing is controlled by the presence (or absence) of particular chemical groups attached to histone proteins that serve to pull the nucleosomes closer together or allow the structure to be more open.
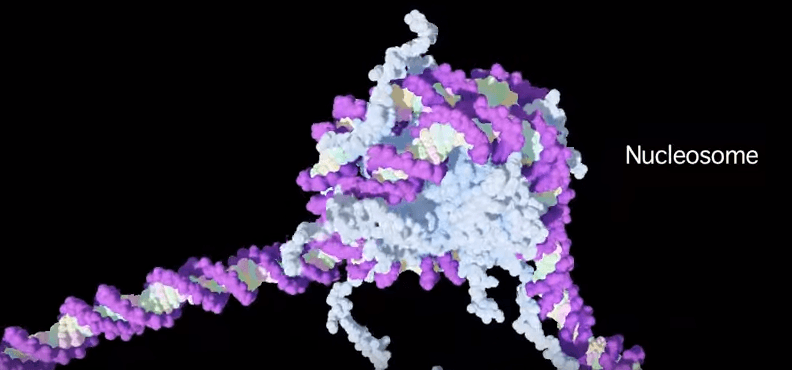
For example, when a molecule called an acetyl group (COCH₃) attaches to a particular part of a histone, it causes the histone structure to relax, and the nucleosomes are further apart. The histone/nucleosome structure is ‘open’. It’s more easily accessed by the transcription machinery that reads the genes to generate messenger RNA, which is then used to build the proteins that make the cells of the organism—the gene is active, or ‘turned on’.
However, other chemicals, such as methyl groups (CH3) can serve to pull the histone structure closer together (but only in some cases, depending on where exactly the methyl group attaches itself!), packing the nucleosome beads on the necklace very tightly together. This means the DNA can’t be accessed. Any genes contained in those sections of the DNA won’t be read—they are inactive, or ‘turned off’.
There is no evidence to date that this sort of epigenetic marking can be passed on from parent to offspring.
DNA methylation
A methyl group can also attach to the DNA double helix with a different purpose to histone spacing regulation. It bonds with the cytosine base (cytosine’s chemical structure means that it is the only base that methyl groups can buddy up with) mainly in CpG dinucleotides (a section of DNA where a cytosine base and guanine base are connected by a phosphate molecule). This is known as DNA methylation.
There are certain regulatory markers within the DNA sequence that precede the beginning of a particular gene, known as the ‘promoter region’ for that gene. Around 60 per cent of genes contain a ‘cluster’ of multiple CpGs (known as CpG islands) around their promoter region.
This is important, because the majority of genes in normal cells have no methylation in these CpG islands. When CpGs located in gene promoter regions are methylated, the gene is inactivated. The methylation of the gene’s promoter region is part of a process that prevents the messenger RNA for the gene from being produced. Generation of messenger RNA is the first step in transcription so if this is hindered, the DNA does not get expressed: the gene is silenced.

Interestingly, apart from the promoter CpG islands and other regulatory regions, the rest of the genome is heavily methylated. This is thought to play a role in maintaining the genome’s structure and stability. Some of the genome’s DNA sequences contain what are known as retrotransposons—replicated chunks of DNA that have incorporated themselves into the genome over time, a bit like a copy and paste function that replicates DNA that isn’t really needed. To keep this extraneous information quiet, much of it is methylated throughout the genome.
Inheritance of DNA methylation
Generally, the DNA methylation pattern across our genome is a stable, lasting epigenetic marker, set early in development.
As new cells are produced, methylation patterns are replicated by the actions of an enzyme called DNA methyltransferase 1. During DNA replication, the DNA strand splits into two, and one of these, called the mother strand, is ‘read’ by the replication machinery and used as a template to build the complementary second strand. After the DNA has been replicated, the newly-synthesised strand is ‘checked’ by the DNA methyltransferase 1 enzyme. It can recognise locations on the mother DNA strand that have methylation, and will attach a methyl group to the appropriate cytosine base on the newly-synthesised strand.
It’s this process that makes for the replication and stability of DNA methylation as an epigenetic marker during the course of an individual’s lifetime.
However, there are two important moments where the DNA methylation slate is essentially cleared. The first is upon fertilisation and the first few rounds of cell division where the DNA methylation marks are removed from the egg and sperm DNA. In this very early stage of development, all the cells contained within the developing embryo (called a blastocyst at this stage of development) are toti-potent cells. This means that they are stem cells that have the potential to develop into any type of cell, so they cannot contain any sort of epigenetic markers that would hamper this toti-potency.
The second resetting occurs in the early germ cells of the embryo that will go on to become either sperm or eggs, which are present in an embryo during the second week of development.
Not all the methylation is cleared—some of the DNA methylation that occurs throughout the genome is important and necessary to maintain a healthy organism. Imprinted genes (genes from either our mother or father) are marked by differential methylation patterns. It’s important that only one copy of these genes are expressed and so the methylation mark is protected from de-methylation.
Sometimes, other DNA methylation patterns are not completely wiped clean—they escape the resetting process and are passed down from parent to offspring. This sort of inheritance is unpredictable and does not follow the ‘normal’ Mendelian pattern of genetic inheritance, so it’s hard to predict epigenetic outcomes.
As for the re-establishment of DNA methylation patterns after the resetting process, this occurs through the action of two methyltransferases (3a and 3b). No one’s quite sure yet exactly what dictates this de novo DNA methylation, which has a unique signature in each cell type.
RNAs
RNA is similar to DNA except it has a slightly different chemical make-up and is generally found as a single-stranded conformation, rather than double stranded. There are quite a few different types of RNA and they can perform a number of genetic processing functions. In the epigenetic context, there are some RNAs that can influence gene expression directly, or they can also play a role in some of the processes of histone modification and DNA methylation.
Small non-coding RNAs
There are several types of small non-coding RNAs. Two of these, microRNAs and short interfering RNAs (both around 21–22 nucleotides long) can prevent the transcription of genes in two ways. Firstly, they can bind to the messenger RNA to make a double-stranded RNA sequence. Because some viruses are made from double-stranded RNA, the cells jump into defence mode, and act to destroy the RNA sequence.
Alternatively, if the RNAs pair up with the messenger RNA imperfectly they simply act to inhibit the construction of proteins on the messenger RNA, blocking further processing of the gene.
Long non-coding RNAs
These are capable of regulating gene expression by influencing and modifying the production of messenger RNA. Long non-coding RNAs contribute to gene-silencing, heritability of epigenetic marks, stem cell differentiation, immune system regulation and cancer metastasis.
Epigenetic outcomes
One of the most significant epigenetic outcomes is cell differentiation. However, it’s not the only one.
Because epigenetic markers act to control and regulate the activity of our genes, our ‘epigenome’ can influence our physical appearance in much the same way as our genes do, but with no change to the underlying genetic code.
X inactivation
Another important epigenetic process is X inactivation. A human’s genes are grouped in clumps we call chromosomes, and we each have 46: 23 from our mother and 23 from our father. Two important chromosomes are the X and Y, as they are what determine a person’s sex. A female will have two XX chromosomes, while males have one X chromosome, and also a Y chromosome. But we only really need the information contained on one of these chromosomes: males need the Y information, while females use the information on the X chromosome. But every female has two X chromosomes, so double the amount of information required. What to do with all that information on the extra Xs? Keep it quiet.
The epigenetic mechanism of X inactivation is responsible for silencing one of the two X chromosomes in every cell of a female’s body, so that each cell has just one activated X chromosome. The curious thing is, for any given gene, it can be either the maternal OR paternal copy of the X chromosome that is activated—giving rise to a characteristic known as mosaicism. The particular genes on the maternal X chromosome may be different variations (alleles) to those on the paternal X chromosome, so the individual ends up displaying a bit of each variation. Some cells will have the paternal X chromosome activated, while other cells have the maternal X chromosome activated.
This is illustrated by the appearance of calico cats. This cat has a distinctive mottled (tortoiseshell) fur colour, but it’s only ever present on the females. This is because the genes for fur colour are on the X chromosome, and as the female cat receives X chromosomes from both her mother and father it can be either the maternal or paternal chromosome that’s active in the different hair cells over her body.
X inactivation is done through a combination of histone modification, DNA methylation and the action of long non-coding RNAs. Xist is a long non-coding RNA produced by the X chromosome that will become inactive. The other X chromosome, that will become active, produces a different long non-coding RNA called TSix (the opposite of Xist!).
When things go wrong—epigenetics and disease
As an epigenetic marker can control the activity of a particular gene, it mimics genetic disease. This is the functional equivalent of a genetic mutation, even though the underlying genetic code may be completely ‘normal’.
Cancer is the result of uncontrolled cell growth and multiplication, and there are three types of known cancer-causing genes: oncogenes (tumour-genes), tumour suppressor genes and DNA repair genes.
However, cancer is now recognised as both a genetic and epigenetic disease. For one thing, there are far too few genes that are known to be implicated in cancer to account for the widespread occurrence of the disease—there has to be additional factors.
There are also cases in which people are suspected to have a genetic predisposition to certain types of cancer (perhaps due to family history), but with no apparent evidence of mutated genes. Now that whole genome sequencing has become so much simpler, mutations are being found in the regulatory regions of the genome.
In other cancer cases, the mutation is in a gene coding for a protein that is critical to the epigenetic machinery, such as DNA methyltransferases, DNA demethylases and histone modification enzymes.
The DNA in cancer cells often has a radically different methylation pattern to those of normal healthy cells. In a healthy cell, the promoter region of genes are generally not methylated, while the rest of the genome is heavily methylated. In some cancer cells, this methylation pattern gets messed up: promoter regions are heavily methylated and the non-gene-coding part of the DNA is demethylated. The end result is entire regions of the genome being abnormally suppressed or active.
There are several other common diseases, such as cardiovascular disease, diabetes and other auto-immune diseases that show evidence of some sort of non-Mendelian inheritance between generations of a family. Scientists are intrigued as to whether these are in fact epigenetic signatures, or genetic changes in genes that code for proteins integral to the epigenetic machinery.
If we can better understand how epigenetic diseases work, it offers great hope for future treatments—the technology to fix mutated DNA is still in its infancy, but we can possibly fix epigenetic errors with drugs that can cause demethylation or changes to histone modifications.
Environmental influences on the epigenome
This is where epigenetics gets really interesting. It’s been found that external influences such as diet, stress and exposure to things like cigarette smoke, heavy metals or pesticides may potentially affect DNA methylation patterns, leading to epigenetic imprints or effects that are caused by a person’s environment or lifestyle. Studies have found that young identical twins have nearly identical epigenomes, but older twin pairs have striking differences in their DNA methylation patterns.
What’s more, DNA methylation patterns are laid down during development. This means the environment or diet of a pregnant woman can potentially have subtle effects upon the methylation patterns laid down in the epigenome of her developing foetus.
To maintain normal DNA methylation patterns (not to mention adequate DNA synthesis and healthy development), we need a number of nutrients from our diet, including a source of methyl groups such as methionine (an amino acid) or choline (an ammonium-based molecule, usually grouped with the B-complex vitamins) and folate (folic acid, found in leafy greens, legumes and oranges). Although folate is not itself a methyl donor, it interacts with methionine and choline to ensure adequate DNA methylation throughout the genome.
What’s more, as a foetus develops, it’s growing its own germ cells—the cells that will go on to become either its sperm or eggs—thus setting the epigenome of its own future offspring. This means that the environmental influences experienced by a pregnant woman can potentially affect not only her own children, but also their children… in effect, she’s pregnant not only with her child, but also her grandkids.
Can your grandma's diet affect you?How could your grandma's diet while pregnant have an affect on generations down the line? This highlight from The Victor Chang Cardiac Research Institute's Associate Professor Cath Suter's fascinating talk on epigenetics -- how and why genes switch on or off -- explains. You can watch the whole talk here:https://www.youtube.com/watch?v=KpWQXSvZPWg Sign up to our events list to keep up to date with Academy events and talks:https://www.science.org.au/subscribe-academys-newslettersSee all up-coming Academy events and public talks here:https://www.science.org.au/events
Posted by Australian Academy of Science on Monday, November 16, 2015
Epigenetic emotions
There are other studies that have found extremely interesting potential epigenetic impacts upon our emotional wellbeing. However, these studies need to be carefully interpreted. Many of them are conducted on animal models and it is unclear if they can be extrapolated to humans.
Other (fairly small scale) studies have claimed that the traumas suffered by one generation can leave epigenetic markers that are passed down through the generations—things like an inherited response to stress in the children of Holocaust survivors, and lower cortisol levels in the children of women who were pregnant at the time of the September 11 terrorist attacks and developed post-traumatic stress disorder as a result of the attacks. Although this idea is intriguing and appealing on various levels, we must be careful to not overblow these findings and apply them in a broad brushstroke manner, especially before more studies have provided further evidence. Many established scientists remain sceptical.
Some of the most compelling experiments examining emotional epigenetics effects looked at the mothering habits of rats. They found that the babies (or pups) of attentive mothers that licked and groomed their pups more often grew up to be ‘happier’ than the pups of inattentive mothers. Studies of the different rats’ brains showed that the inattentive mothers’ pups had high methylation levels on the gene that controlled the production of glucocorticoid receptors, which control sensitivity to stress hormones. The attentive mothers’ pups’ glucocorticoid receptor genes were not methylated, meaning more glucocorticoid receptors were expressed and the pups were more sensitive to glucocorticoid.

When attentive mothers adopted the pups of inattentive mothers, the rats grew up happier, suggesting that the result was not due to an underlying genetic difference. To further clinch their hypothesis, the researchers gave pups of inattentive mothers a dose of a drug that removes methyl groups, which resulted in these rats not showing the negative impacts of the poor mothering, and their brains free of the epigenetic methylation.
This was because of the epigenetic influence of the mothering behaviour upon the baby rats at a crucial time of their development—shortly after birth is when their hypothalamic-pituitary-adrenal (HPA) axis develops. This is the hormonal pathway that controls the glucocorticoid receptors.
As our HPA axis is fully developed by the time we are born, we can’t really extrapolate these results directly to humans. It’s easy to study rats; not so easy to properly study humans. It’s also easy to overstate the results of some these types of epigenetic discoveries, and being unable to truly manipulate the human epigenome as you can in the lab with a mouse or rat, it’s difficult to really nail down solid conclusions about the potential to pass down emotional epigenetic markers through the generations.
While it’s hard to pin down exactly the role epigenetics plays in inheriting emotional responses, it’s clear our genes do not contain our entire destiny. The additional layer of instructions provided by epigenetic marks is critical to the regulation and control of our genetic information, with implications not only in basic cell development and differentiation, but also disease and other physical conditions.