Scientists have been observing examples of biased inheritance generated by natural gene drive mechanisms for many years. The concept of a ‘synthetic gene drive’ was devised almost 50 years ago by Christopher Curtis who proposed using translocations (rearrangements of genetic material) to drive anti-pathogenic genes into wild species (Curtis, 1968). This idea was further developed by Austin Burt (2003; 2014), an evolutionary geneticist, who discussed how a synthetic gene drive could be used to prevent insects spreading diseases such as malaria.
There are many different types of natural gene drive mechanisms (Appendix 1). These can be characterised by attributes such as the rate of spread, species specificity, fitness cost, susceptibility to resistance, removability and reversibility (Champer et al., 2016). The rate of spread is an important consideration. So called ‘high threshold’ gene drives would only spread if the number of individuals with the drive genotype reaches a high level. These types of drive systems could be confined to local areas and breeding populations by controlling the number of individuals with and without the drive. In contrast, ‘low threshold’ gene drives, which are considered invasive, would spread with a low initial release, requiring only a small number of gene drive-carrying organisms to be released to spread. Natural Wolbachia infections provide examples of drives with high and low thresholds (Nguyen et al., 2014; Hoffmann et al., 2011). It is worth noting that no synthetic gene drives have yet been released into wild populations so the concepts discussed here are untested to date on such systems.
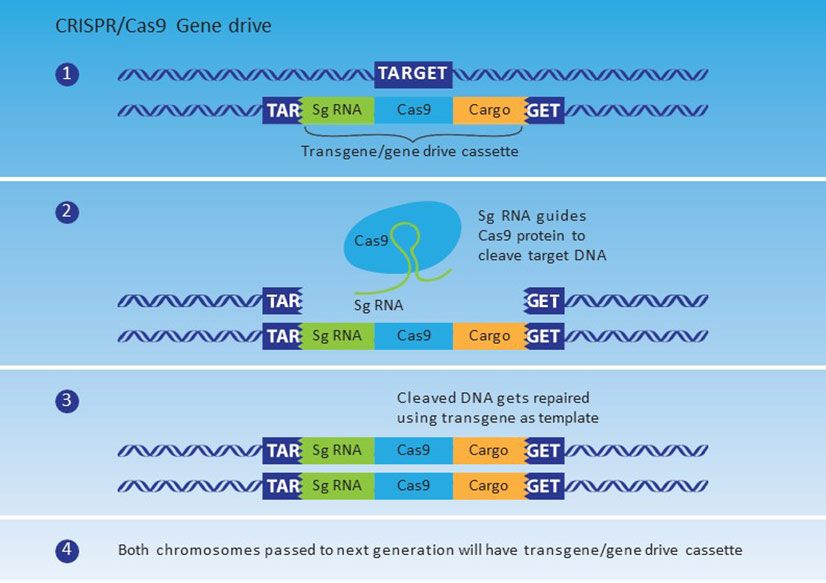
Recent advances in gene editing tools allow organisms to be edited much more efficiently and more accurately than previously possible. Scientists can now harness gene drive mechanisms which were previously merely theoretical to control or alter natural populations. While not a gene drive tool in its own right, clustered regularly interspaced short palindromic repeats of base sequences (CRISPR), can be used as part of a system to produce a synthetic gene drive. When CRISPR is paired with a guide RNA and with specific proteins, such as Cas9 (CRISPR associated protein 9) that cuts DNA, it can be used to efficiently edit genetic material. In natural prokaryotic systems, CRISPR/Cas9 is produced by host bacteria to remove viral DNA by targeting repeats associated with viral insertions, as a kind of immune system to combat infections. For gene editing purposes, the Cas9 protein and guide RNA are injected into the cell to cut the DNA at a sequence complementary to the RNA guide. For synthetic gene drives, the target organism is transformed with a construct that includes the gene for the Cas9 protein, a guide RNA that is complementary to the sequence at the insertion site, and the ‘cargo’ gene controlling the desired trait (Figure 2). The guide RNA directs Cas9 to produce a double stranded cut in the DNA at the target site in the other chromosome. This triggers the cell’s repair mechanism, which copies the entire construct (Figure 2). If germ cells are targeted, the new sequence can then be passed on to offspring ensuring the editing changes can occur in each generation. A CRISPR-based gene editing technique was used in all four synthetic gene drive proof-of-concept studies in 2015. These studies generated laboratory-based gene drives in yeast Saccharomyces cerevisiae (DiCarlo et al., 2015), fruit fly Drosophila melanogaster (Gantz & Bier, 2015) and two mosquito species Anopheles stephensi (Gantz et al., 2015) and Anopheles gambiae (Hammond et al., 2016).
© 2026 Australian Academy of Science